| Accession: | |
|---|---|
| Functional site class: | Atg8 protein family ligands |
| Functional site description: | The autophagy-related protein Atg8 and its homologues LC3 and GABARAP play an important role in selective autophagy. During autophagy, Atg8 proteins get directly conjugated to phosphatidylethanolamine (PE) lipids to mediate membrane fusion events involved in autophagosome biogenesis such as phagophore formation and elongation. In addition, different Atg8 protein family members can recruit specific adaptors bound to ubiquitylated proteins, organelles or pathogens for degradation. Many of these adaptor proteins contain an LC3-interacting region (LIR) that mediates binding to Atg8 and Atg8-related proteins. These LIR:Atg8/LC3/GABARAP interactions are essential for cellular cell homeostasis as well as the control of intra- and extracellular stress conditions. |
| ELMs with same func. site: | LIG_LIR_Apic_2 LIG_LIR_Gen_1 LIG_LIR_LC3C_4 LIG_LIR_Nem_3 |
| ELM Description: | The core of the LIR motif is defined by four amino acids and adopts a β-strand conformation that binds by β-augmentation, forming an intermolecular parallel β-sheet with the second β-strand of Atg8 protein family members (Rogov,2014). There is an absolute requirement for an aromatic residue at the N-terminal side of the LIR core and a large, hydrophobic residue at the C-terminal side. Structural studies have revealed that the side chain of the aromatic residue of the LIR motif binds deeply in HP1 whereas the hydrophobic residue docks to HP2 (2ZJD; Ichimura,2008). Position +2 in the core is solvent accessible and aromatic residues are not favoured. The presence of positive charges in the binding domain also restricts the +2 position to not-positively charged residues. The fixed distance from HP1 to HP2 makes it inadequate for a tiny residue or a Pro at +2 and +3 positions. The core motif is generally preceded by a varying number of acidic residues or by Ser or Thr residues that can be phosphorylated to incorporate a negative charge. These residues commonly occur within three positions N-terminal to the core motif. The negative charge of these acidic or phosphorylated residues has been shown to strengthen the LIR:Atg8/LC3/GABARAP interaction (Rogov,2013). Additional acidic residues or Ser/Thr phosphorylation sites that strengthen the interaction are sometimes observed in the positions between the aromatic (+1) and hydrophobic (+4) residue. A Trp residue is energetically favoured for this interaction over a Tyr or Phe residue, but the lower binding affinity can be compensated by electrostatic interactions between acidic residues or Ser/Thr phosphorylation sites of the LIR motif and basic residues in the N-terminal arm of the Atg8 homologues (Wild,2013). Analysis of current known structures indicates that the motif can be located at the C-terminus of the protein or be followed by a particular range of acidic or not residues immediately, or up to four positions, after the core. |
| Pattern: | [EDST].{0,2}[WFY][^RKPGWFY][^PG][ILVFM]((.{0,4}[PLAFIVMY])|($)|(.{0,3}[ED])) |
| Pattern Probability: | 0.0036312 |
| Present in taxon: | Eukaryota |
| Not represented in taxon: | Nematoda |
| Interaction Domain: |
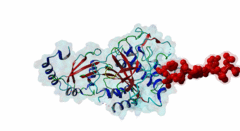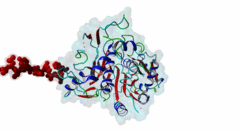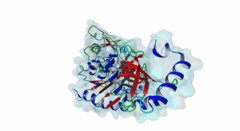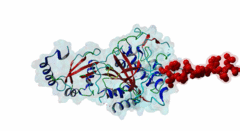
Atg8 (PF02991)
Autophagy protein Atg8 ubiquitin like
(Stochiometry: 1 : 1)
|
Macroautophagy is an evolutionary conserved degradation process that targets macromolecules, organelles and intracellular pathogens and is of vital importance for cellular homeostasis. Many of the proteins involved in autophagy were first identified and named in yeast. In this process, a double membrane structure called the phagophore forms and expands to form a double-membrane vesicle, the autophagosome. Autophagosomes then sequester cargo and eventually fuse with the vacuole in yeast or the lysosome in higher eukaryotes in order to degrade their content (Mizushima,2011). Several autophagy-related proteins (Atgs) are required for the formation of the autophagosome and they are highly conserved in Eukaryotes. Among these Atg proteins are the yeast autophagy protein Atg8 and its vertebrate homologues which are ubiquitously expressed in all tissues. An upregulation of Atg8 proteins can be observed under various stress conditions (Shpilka,2011). After its translation, the carboxy-terminal region of Atg8 is cleaved in order to expose a glycine residue. Atg8 is then processed by an ubiquitin-like conjugation machinery, which directly conjugates it via its exposed glycine to a PE (phosphatidylethanolamine) lipid. This enables Atg8 to be involved both in cargo recruitment into autophagosomes and the formation and elongation of the autophagosome (Johansen,2011). In contrast to yeast and other fungal species, which have only a single Atg8 protein (P38182), multicellular animals, plants and some protists have several Atg8 homologues. These can be grouped into two subfamilies: the microtubule-associated protein 1 light-chain 3 (MAP1LC3 or LC3) with its paralogous variants LC3A (Q9H492), LC3B (Q9GZQ8) and LC3C (Q9BXW4), with two spliced isoforms of LC3A, and the γ-aminobutyrate receptor-associated paralogous proteins including GABARAP (O95166), GABARAPL-1 (Q9H0R8) and GABARAPL-2 (P60520). Proteins binding to the Atg8 protein family typically possess a short hydrophobic LC3-Interacting Region (LIR) motif, which is often also referred to as Atg8-family Interacting Motif (AIM) in yeast. The LIR is required for these proteins to bind Atg8 and its homologues. Proteins containing LIR motifs include cargo receptors, members of the basal autophagy apparatus, proteins associated with vesicles and of their transport, Rab GTPase-activating proteins (GAPs) and specific signalling proteins that are degraded by selective autophagy. They represent an essential part of autophagosome formation, transport and maturation (Birgisdottir,2013). Proteins belonging to the Atg8 family have a C-terminal domain (PF02991) belonging to the large ubiquitin-like (UBL) domain superclass which consists of a four-stranded β-sheet wrapped around a central α-helix. The hydrophobic residues of the central α-helix and β-strand 2 of the UBL core form a hydrophobic pocket (HP2). Preceding and extending the UBL core is an N-terminal arm with two α-helices. This N-terminal subdomain varies among the different Atg8 subfamilies. It is packed onto the core UBL and forms another deep hydrophobic pocket (HP1) (2KWC; Kumeta,2010). The LIR docking site is located at the interface of the UBL core and the N-terminal arm and consists of the two hydrophobic pockets HP1 and HP2. When cells are invaded by pathogenic bacteria, autophagy is used to capture and subsequently eliminate the invader in conjunction with the lysosome: this process is also called xenophagy. Pathogens have been found to counteract this process by evolving mechanisms to evade or abuse autophagy. Legionella pneumophila secretes the protease RavZ to irreversibly deconjugate LC3-PE by cleaving the Glycine-containing C-terminal region. In order to recognize its target protein, RavZ uses three mimics of the LIR motif to bind to up to two LC3 molecules (Kwon,2017). A secreted protein from the oomycete Phytophthora infestans binds to the autophagy-related protein ATG8CL from potato outcompeting with the host cargo receptor affecting the plant defense (Dagdas,2016). Viruses also interact with the autophagy system. Influenza A virus requires the presence of LC3 at the plasma membrane during virus budding to form filamentous virions and, ultimately, for enhanced virion stability. The relocalization of LC3 depends on a LIR motif in the cytoplasmic tail of Matrix 2 ion-channel protein (Beale,2014). Coronavirus, including SARS-CoV-2, proteins accumulate or prevent the efficient formation of autophagosomes (Koepke,2021): However, at time of writing, no LIR motif has been identified in coronavirus proteins. LIR motifs described in nematodes are less specific, as they accept a wider range of residues after the aromatic residue including tiny residues at the +2 position and Tyr at +4 of the core. Likewise, Apicomplexa LIRs use a Pro residue at the +4 position which is not observed in other eukaryotes. For these reasons, two non-canonical classes exist: LIG_LIR_Apic_2 and LIG_LIR_Nem_3. A structural difference in LC3C homologues reduces the exposure of HP2 compared to other Atg8 homologues, this restricts the core to a 3-amino acid core and is described in the ELM class LIG_LIR_LC3C_4. |
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement