| Accession: | |
|---|---|
| Functional site class: | PAM2 motif |
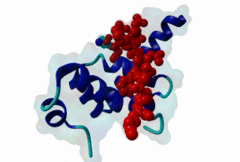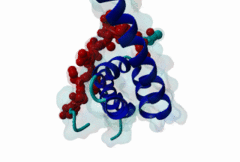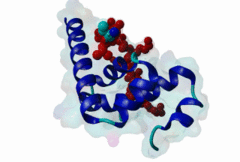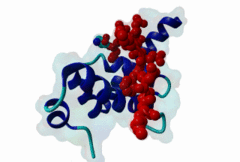
| Functional site description: | The PABP-interacting motif 2 (PAM2) mediates binding of proteins to the MLLE/PABC peptide-binding domain found in poly(A)-binding proteins and HYD E3 ubiquitin ligases. There are two variants of the PAM2 motif that bind to overlapping but distinct sites on the MLLE domain, adopt a different orientation at their termini and consequently have different recognition elements in these regions. While the core region around a critical phenylalanine is similar in the LIG_PAM2_1 and LIG_PAM2_2 variants and makes identical contacts with the MLLE domain, the N-terminal sequence in the LIG_PAM2_1 provides a hydrophobic residue that is essential for efficient binding and is lacking in LIG_PAM2_2 motifs, while the C-terminal part of the LIG_PAM2_2 variant makes a turn and follows an alternative path on the MLLE surface that depends on an aromatic residue that is absent from LIG_PAM2_1 motifs. |
| ELMs with same func. site: | LIG_PAM2_1 LIG_PAM2_2 |
| ELM Description: | Binding of the LIG_PAM2_1 mainly involves hydrophobic interactions, the most important of which are mediated by the residues in position 3 (lacking in LIG_PAM2_2) and 10 (corresponds to position 5 in LIG_PAM2_2). The residues in positions 1 and 2 of the motif, most often polar or charged residues, do not participate in binding, except for eRF3, which contains two overlapping motifs, where phenylalanine in position 1 of the C-terminal motif bends back to bind in the hydrophobic pocket that is normally occupied by the hydrophobic residue at position 3, which is most frequently leucine, but in some cases phenylalanine or proline. The side chain of the asparagine or serine residue in position 4 is involved in an intermolecular salt bridge and forms an intramolecular hydrogen bond with the amide of the residue in position 6. The latter interaction stabilizes the beta-turn conformation of the peptide. The regular occurrence of proline in position 5 might be due to its propensity to form such a beta-turn. Position 7 is invariantly alanine, which is involved in different hydrophobic interactions with multiple MLLE residues. Introduction of a bulky residue at this position was found to decrease the affinity for the peptide. Position 10 is the single most important residue for binding and in most cases it is occupied by phenylalanine, but exceptionally by tryptophan in LARP4 and LARP4B, or possibly also by tyrosine in the Arabidopsis thaliana proteins CID5 and CID6. Since CID5 is the only example of an experimentally validated motif with a tyrosine in position 10, this feature might be specific for plants. The residue in position 10 occupies a hydrophobic pocket between helices 2 and 3 and is the major binding determinant. In case of a phenylalanine in this position, a hydrophobic residue is found in position 12, with a clear preference for proline. AtCID5 and AtCID6 also contain a proline at position 12. For Fungi and some TAXON:7215 sequences, the regular expression is less strict. |
| Pattern: | ..[LFP][NS][PIVTAFL].A..(([FY].[PYLF])|(W..)). |
| Pattern Probability: | 0.0000100 |
| Present in taxon: | Eukaryota |
| Interaction Domain: |
PABP (PF00658)
Poly-adenylate binding protein, unique domain
(Stochiometry: 1 : 1)
|
The PABP-interacting motif 2 (PAM2) directly binds to the MLLE/PABC domain that is found in poly(A)-binding proteins (PABP) like PABPC1 and in members of the HECT domain-containing Hyperplastic Discs (HYD) protein family of E3 ubiquitin ligases (Albrecht,2004, Lim,2006). This domain consists of a conserved bundle of five alpha-helices, of which the N-terminal helix is lacking in some proteins. The interaction with PAM2 sequences involves the most conserved helices 2, 3 and 5, with the exception of yeast, where only helices 2 and 3 mediate binding. PABPC1 binds to the 3’-poly(A) tail of mRNA molecules via four RNA recognition motifs (RRMs) and recruits different regulatory proteins, which modulate translational activity and mRNA stability, by binding to their PAM2 motifs via its C-terminal MLLE domain (Siddiqui,2007). LIG_PAM2_1 motif-containing PABPC1-binding proteins include Paip1 and Paip2, which stimulate or repress translation by stabilizing or destabilizing, respectively, the closed loop structure of mRNA that is formed by the interaction between PABP and the translation initiation factor eIF4G. Other examples are the eukaryotic release factor eRF3, which contains two overlapping LIG_PAM2_1 motifs, and Tob1 and Pan3, which recruit the deadenylase complexes Caf1-Ccr4 and Pan2-Pan3, respectively, to the mRNA poly(A) tails. These complexes are involved in translation-dependent, eRF3-mediated mRNA decay and translation termination. Tob1 and its family member Tob2 contain two distinct LIG_PAM2_1 motifs, and evidence indicates that in both cases the C-terminal motif is the main interaction site for binding to PABP. PABPC1 binding partners containing the LIG_PAM2_2 motif variant are so far restricted to animal-specific GW182 family proteins, which are part of the RNA-induced silencing complex (RISC) and are essential for microRNA (miRNA)-mediated gene silencing (Jinek,2010). Silencing is achieved by inhibiting translation and promoting degradation of mRNA, and depends on binding of GW182 proteins in the miRNA-loaded RISC to PABPC1. Additional, possibly indirect, interactions also occur between the RRMs of PABPC1 and the C-terminal region of GW182 family members. While in human GW182 proteins PABPC1 binding is predominantly mediated by their PAM2 sequences, in D. melanogaster GW182 the motif, which binds with lower affinity, also contributes to but is dispensable for this interaction in immunoprecipitation experiments (Huntzinger,2010). The tumor suppressor protein HYD (EDD/UBR5) belongs to the family of HECT domain-containing E3 ubiquitin ligases, which target specific proteins for proteasome-dependent degradation. Evidence suggests that HYD is involved in proliferation and DNA damage signaling. Little is known about the function of the MLLE-PAM2 interaction in HYD E3 ligases, but it has been suggested that PAM2-containing proteins may be targeted for ubiquitination by HYD through binding to its MLLE domain, which is positioned directly adjacent to the catalytic HECT domain. So far, all known PAM2-mediated interactions with HYD involve instances of the LIG_PAM2_1 variant. The structural features of PAM2 binding to the MLLE domain of PABP or HYD are very similar, and similar specificity and binding affinity has been demonstrated for various peptides (Lim,2006). |
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement