| Accession: | |
|---|---|
| Functional site class: | FFAT motif |
| Functional site description: | The FFAT motif binds to the Major Sperm Protein (MSP) domain of the integral Endoplasmic Reticulum (ER) membrane proteins Vesicle Associated Membrane Protein (VAMP) Associated Proteins, VAP-A and VAP-B. This motif was initially linked to the targeting of lipid trafficking and lipid sensing proteins to the cytosolic face of the ER. Further studies indicate that VAP-FFAT binding plays a part in the formation of membrane contact sites between the ER and other cellular membranes. Most known instances of FFAT motifs occur in mammal and yeast proteins but additional examples have been reported in pathogenic bacterial and viral proteins. VAP-FFAT interactions may play a role in disease etiology, as mutations in VAP-B have been implicated in late onset Spinal Muscular Atrophy and Amyotrophic Lateral Sclerosis – type 8. Another protein, MOSPD2, which also has an MSP domain has been identified as a second binding partner for FFAT proteins. |
| ELMs with same func. site: | TRG_ER_FFAT_1 TRG_ER_FFAT_2 |
| ELM Description: | The FFAT motif was initially identified as a short sequence shared between a set of ER-targeted proteins, that was shown to mediate interactions with yeast members of the VAP protein family (Loewen,2003). The consensus core motif was described as EFFDAxE. This core motif is supplemented by a seventh element, the flanking regions (mainly upstream) which contain multiple acidic residues. The acidic tract seems to be crucial for the initial, non-specific binding to the positive surface of the VAP proteins which is further amplified by key residues in the core motif, particularly the residues in the 2nd and 5th positions. After the original description of the motif, many instances have been reported with various alterations. The second position is the most conserved, tolerating only a phenylalanine or a tyrosine. The aromatic ring of these amino acids binds in a hydrophobic pocket created by the aliphatic side chains of critical VAP residues, including Lys87 and Met89. A second hydrophobic pocket binds the 5th residue of the motif, typically an alanine or a cysteine. Position 3 of the core motif is rather variable, allowing for a number of substitutions. Aside from the FFAT motif binding with the surface of VAP proteins, a second interaction is revealed in the crystal structure. Two FFAT motifs bind to each other, revealing a “double sandwich” conformation. This bond is mediated by the side chain of the aspartic acid residue in the 4th position of the motif which forms a symmetry related hydrogen bond with the backbone of the aspartic acid of the second FFAT motif. Originally, this position was thought to not tolerate any substitutions but one instance with a glutamic acid replacing the aspartic acid has proven to be functional (Hantan,2014). Additionally, an acidic amino acid is required in the first position. In the structure, this residue seems to interact with the second VAP protein of the dimer. |
| Pattern: | [EDS].{0,4}[ED][FY][FYKREM][DE][AC].{1,2}[EDST] |
| Pattern Probability: | 0.0000103 |
| Present in taxons: | Homo sapiens Opisthokonta Rattus norvegicus Saccharomyces cerevisiae Viruses |
| Interaction Domain: |
Motile_Sperm (PF00635)
MSP (Major sperm protein) domain
(Stochiometry: 1 : 1)
|
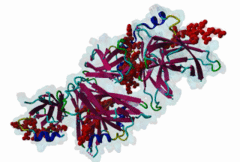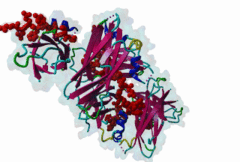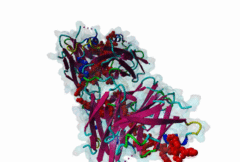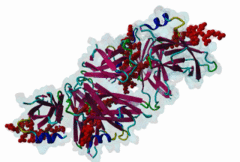
The Endoplasmic Reticulum (ER) is a dynamic structure that serves as a site of bulk membrane lipid biogenesis for most of the cell organelles with a major portion of lipid production being associated with the cytosolic leaflet of the ER membrane (Baumann,2001). Thus, many proteins with roles in lipid metabolism must access the ER surface. VAP proteins are ER membrane proteins that are highly conserved among eukaryotes, consisting of a C-terminal transmembrane domain, a flexible linker that may contain a short coiled coil, and a cytosolic N-terminal MSP domain (Lev,2008, PF00635). VAPs contribute to the targeting of proteins to the ER through the interaction that occurs between their MSP domain and the FFAT motif of various proteins in both mammals and yeast (Loewen,2003). Many of the proteins that contain FFAT motifs function in lipid sensing and lipid exchange. Additionally, a role of VAPs in the formation of Membrane Contact Sites (MCSs) has emerged. MCSs are distinct domains between the ER and other organelles where organellar membranes are bridged by proteins (often with a gap less than 30 nm), but do not fuse. MCSs have been proposed to facilitate the non-vesicular trafficking of small molecules, such as lipids (Helle,2013). VAP-FFAT interactions have been shown to control the formation of MCSs between the ER and other cellular membranes, including the plasma membrane, mitochondria, late endosomes, peroxisomes and vesicles that contain uptaken, possibly pathogenic, bacteria. The 3D structure of the MSP domain-FFAT motif (rat VAP-A with rat ORP1) interaction has been solved using X-ray crystallography (1Z9O). This interaction is mediated by a positive patch on the MSP domain surface, which is strongly conserved (e.g. in Saccharomyces cerevisiae, Schizosaccharomyces pombe, Arabidopsis thaliana, Caenorhabditis elegans and Homo sapiens) in the VAP-family MSP domains, but not in other MSP domains (Loewen,2005). The crystal structure reveals an unusual cooperatively assembled dimer of dimers in a “double peptide sandwich” conformation. FFAT motifs can also bind VAPs as monomers, as was revealed by a more recently obtained solution structure for VAP-A with the FFAT of OSBP (2RR3). |
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement