
PDB-Structure 1H27showing a peptide from ELM classDOC_CYCLIN_RxL_1
COVID-19 SLiM Links
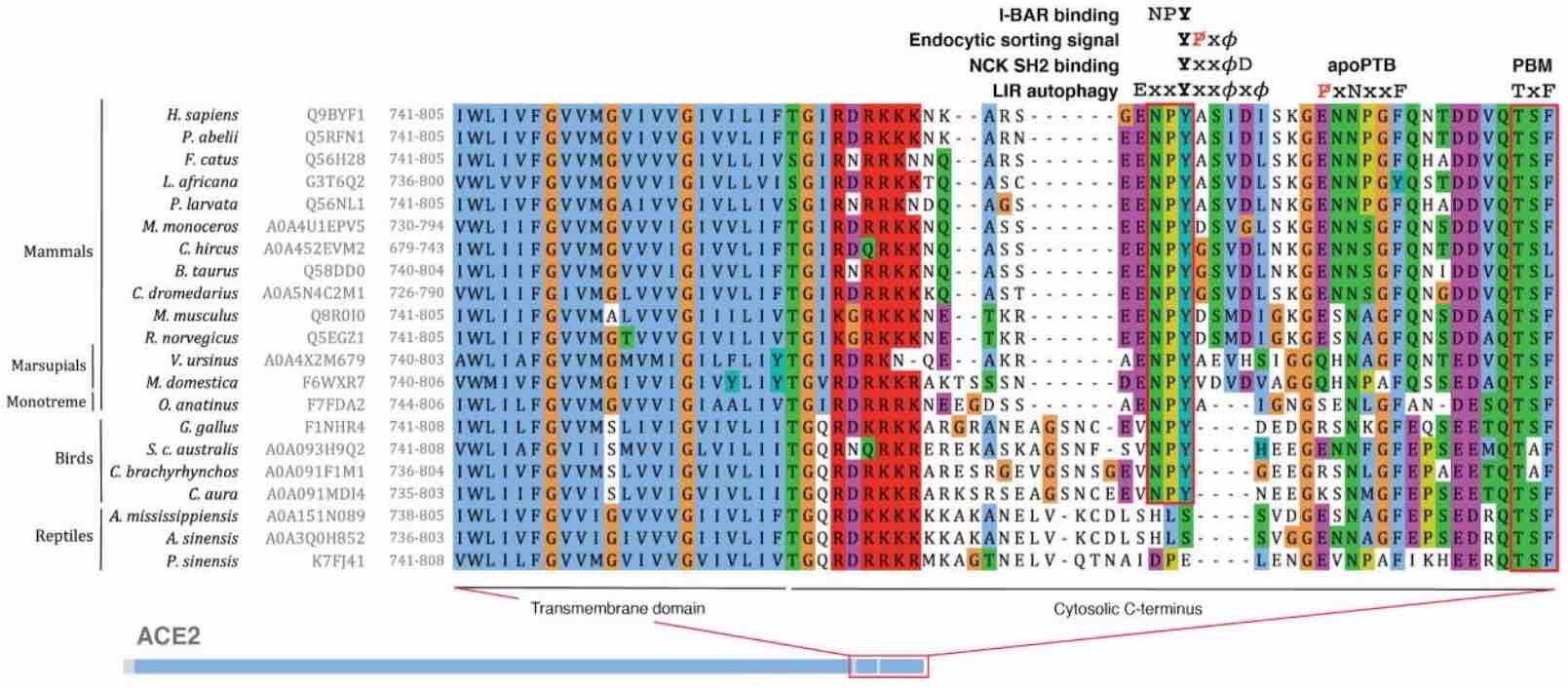
HSTalks interview Gibson and Yamauchi on SARS-CoV-2 entry SLiMs
Our article on SLiM signatures in the SARS-CoV-2 entry system is now available at Science Signaling
Validation experiments for SARS-CoV-2 entry system SLiMs
SPIKE CendR and NRP1 Receptor in SARS-CoV-2 cell entry
Featured Paper
Fast and scalable querying of eukaryotic linear motifs with gget elm
Check out gget elm for computing large-scale queries locally
- ELM database updates
DOC_TBK1_STING_1 motif involved in mediating innate immunity and inflammation has been released.
- ELM database updates
In a huge update, nine new ELM classes
DEG_Cend_DCAF12_1
DEG_Cend_KLHDC2_1
DOC_CYCLIN_RevRxL_6
DOC_PUB_PIM_1
LIG_CtBP_RRT_2
LIG_SH3_PxxxRxxKP_6
LIG_SH3_PxRPPK_7
LIG_SH3_PxxPPRxxK_8
LIG_RBL1_LxSxE_2
have been released. Further, five classes
DOC_PP2B_LxvP_1
DOC_PP2B_PxIxIT_1
LIG_RB_LxCxE_1
LIG_RB_pABgroove_1
LIG_SH3_PxxDY_5
have been substantially revised with ~70 new instances.
- ELM database updates
In this release, three new classes LIG_TRAF3_MATH_PxP_3, LIG_MSH2_SHIPbox_1 and MOD_AAK1BIKe_LxxQxTG_1 have been added. Also, two existing TRAF classes have been comprehensively revised - LIG_TRAF2like_MATH_shPxQ_1 (previously LIG_TRAF2_1) and LIG_TRAF2like_MATH_loPxQ_2 (previously LIG_TRAF2_2).
- ELM database updates
In a major addition seven new classes DEG_Cend_TRIM7_1, DEG_Cend_FEM1B_2, DEG_Cend_FEM1AC_1, LIG_Trf4_IWRxY_1, LIG_MTR4_AIM_1, LIG_IRF7_LxLS_2 and LIG_FERM_MyoX_1 have been added. Three classes, LIG_Nrd1CID_NIM_1, LIG_IRFs_LxIS_1 and LIG_SH2_NCK_1 have been updated.
- ELM database updates
Three new ELM classes LIG_ARS2_EDGEI_1, MOD_LOK_YxT_1 and TRG_NESrev_CRM1_2 have been added. MOD_PIKK_1 has been updated with 18 new instances.
- ELM database updates
Two new classes DEG_CRBN_cyclicCter_1 and TRG_Oom_RxLR_1 have been added to the database.
- ELM database updates
Protein degradation systems got more coverage in ELM. Four new classes LIG_VCP_SHPBox_1, LIG_VCP_VIM_2, LIG_VCP_VBM_3 and DEG_Kelch_KLHL12_1 added in database. In addition, several new instances have been added to DEG_APCC_DBOX_1 entry.
- ELM database updates
ELM now includes more Synapse proteins. New classes LIG_Arc_Nlobe_1 has been added. LIG_CaMK_CASK_1 has been substantially revised and updated to include the latest knowledge.
- ELM database updates
ELM classes MOD_NEK2_1 and MOD_NEK2_2 have been revised and updated to represent stricter and tolerant NEK2 site motif preferences, respectively.
- ELM database updates
Two new classes LIG_LEDGF_IBM_1 and LIG_Menin_MBM1_1 have been added to the database.
- ELM database updates
One new class LIG_TRAF4_MATH_1 added to the database. LIG_TRAF6 has been changed to LIG_TRAF6_MATH_1, the motif has been revised and several new instances added.
Become an ELM sponsor?
The ELM phosphorylation DB
The Switches.ELM DB
Download a movie about a molecular switch involved in the formation of the ALG2/Alix complex
Welcome to the Eukaryotic Linear Motif (ELM) resource
This computational biology resource mainly focuses on annotation and detection of eukaryotic linear motifs (ELMs) by providing both a repository of annotated motif data and an exploratory tool for motif prediction. ELMs, or short linear motifs (SLiMs), are compact protein interaction sites composed of short stretches of adjacent amino acids. They are enriched in intrinsically disordered regions of the proteome and provide a wide range of functionality to proteins (Davey,2011,Van Roey,2014) They play crucial roles in cell regulation and are also of clinical importance, as aberrant SLiM function has been associated with several diseases and SLiM mimics are often used by pathogens to manipulate their hosts' cellular machinery (Davey,2011, Uyar,2014)
ELM Prediction
The ELM prediction tool scans user-submitted protein sequences for matches to the regular expressions defined in ELM. Distinction is made between matches that correspond to experimentally validated motif instances already curated in the ELM database and matches that correspond to putative motifs based on the sequence. Since SLiMs are short and degenerate, overprediction is likely and many putative SLiMs will be false positives. However, predictive power is improved by using additional filters based on contextual information, including taxonomy, cellular compartment, evolutionary conservation and structural features.
ELM DB
The ELM relational database stores different types of data about experimentally validated SLiMs that are manually curated from the literature. ELM instances are classified by motif type, functional site and ELM class. A functional site contains one to many ELM classes, which are described by a regular expression and list experimentally validated motif instances matching this sequence pattern. All data curated in ELM DB can be searched on the ELM website according to the following categories:
- 353 annotated ELM classes
- 4,277 experimentally validated ELM instances in 278 taxons
- 148 ELM methods described in 4,283 articles to experimentally validate ELM instances
- 722 solved PDB structures for curated ELM instances (from PDB)
- 190 globular ELM binding domains (from Pfam, SMART, and InterPro)
- 2,797 interactions mediated by curated ELM instances
- 887 regulatory switches mediated by curated ELM instances (from Switches.ELM DB)
- 1,063 pathways from KEGG involving linear motifs annotated in 1297 Sequences
- 293 viral instances interfering with host cellular processes
- 11 ELM related diseases annotated as being caused by aberrant motif function
- 2 examples where pathogens abuse motifs to deregulate host cells
ELM Candidates
The ELM candidates pages contain lists of candidate classes and instances awaiting curation, and can be extended by users: Anybody can submit candidates, please provide as much information as possible. If you're interested in annotating a full ELM entry or individual instances, get in contact with the ELM team for procedures and awards!
ELM Information
The ELM information pages provide additional details to assist users in searching data in the ELM DB and in searching for putative motifs in query sequences. The Help page explains the use of regular expressions to define sequence patterns of ELM classes and to detect putative motifs in user-submitted query sequences, describes the filters that are applied to increase the reliability of the prediction tool, and defines terms frequently used in the ELM resource. The News lists changes and updates made to ELM, while the Links page lists links to other interesting bioinformatics resources. Funding information, participating groups and a list of ELM-related publications can be found on the About page.
ELM Downloads
Data curated in ELM DB can be downloaded and distributed for non-commercial use according to the ELM Software License Agreement.
ELM API
The ELM prediction tool can also be accessed programmatically via the ELM API. Read the API manual for more information.
Disclaimer
Short patterns applied to proteins are usually not statistically significant: Therefore we can't provide E-values as with BLAST searches. This means that most matches shown are more likely to be false positives than true matches. We hope that ELM server results will prove useful as guides to experimentation but they should not be treated as factual findings.
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
