| Accession: | |
|---|---|
| Functional site class: | NLS classical Nuclear Localization Signals |
| Functional site description: | Many nuclear proteins possess a nuclear localisation signal (NLS) that is recognised by the importer protein importin-alpha. The NLS motif is primarily composed of basic residues and is found in two main variants: a monopartite form and a bipartite form with two short basic segments separated by a flexible linker. Importin-alpha is itself an adaptor for the nuclear transport receptor importin-beta. The latter is docked on the cytosolic side of the nuclear pore via repetitive FG, FxFG and GLFG linear motifs found in several nucleoporin proteins (FG-Nups) (Terry,2009). The cargo loaded importin complexes translocate through the nuclear pore while remaining attached to the flexible FG-Nups. Finally, binding of RanGTP to importin-beta drives cargo release, with the importin-alpha still being bound to nucleoporins located on the nucleoplasmic side. Importin-alpha must be returned to the cytosol to repeat the process. |
| ELMs with same func. site: | TRG_NLS_Bipartite_1 TRG_NLS_MonoCore_2 TRG_NLS_MonoExtC_3 TRG_NLS_MonoExtN_4 |
| ELM Description: | The classical bipartite nuclear localisation signal (bNLS) binds to importin-alpha. It consists of two basic clusters separated by an unconserved linker peptide. The shorter N-terminal basic cluster ("minor NLS site") consists of a pair of basic amino acids (lysine or arginine). The larger, C-terminal basic cluster ("major NLS site") requires three basic amino acids (lysine or arginine) within four positions. The minor interaction is not used for the mNLS which only interacts with the major importin site. For the major NLS site, the core positions are P2-P5. The P2 position is critical to anchor the NLS motif: it must be occupied by a basic residue. Three of the four positions P2-P5 must be occupied by basic residues. Complementary charges and the size of the binding pockets in importin-alpha strictly control the P2 and P3 basic side chain preference: If P2 is arginine, P3 must be lysine; whereas if P2 is lysine, P3 may be arginine or lysine. At least one of P4 and P5 must be a basic residue, provided it is accompanied by a proline or other hydrophobic residue. Acidic residues are not tolerated at P4 and P5. Outside the core motif, there are additional preferences, which, though weaker, clearly play a role in NLS binding affinity, especially at P1, P6 and P7. P1 and P6 both have a preference for basic, Pro and hydrophobic residues. Acidic residues are rejected in P1 and P6, though preferred at P7. The linker region shows only weak amino acid preferences, notably for Pro, Gly and sometimes Ala, that seem to facilitate the nestling of the NLS peptide along the curvature of the importin surface. The length of the linker is almost always within 7-15 residues. The rejection of acidic residues in most positions binding the major site, may allow some NLS activities to be regulated by phosphorylation as Ser and Thr residues are quite often found in the more variable positions as well as in the linker. |
| Pattern: | [KR][KR].{7,15}[^DE]((K[RK])|(RK))(([^DE][KR])|([KR][^DE]))[^DE] |
| Pattern Probability: | 0.0002588 |
| Present in taxon: | Eukaryota |
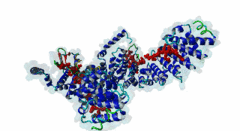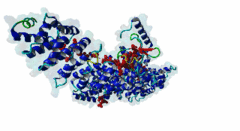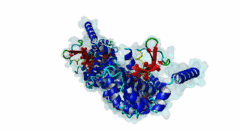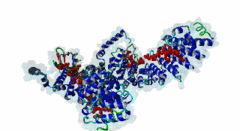
| Interaction Domain: |
Arm (PF00514)
Armadillo/beta-catenin-like repeat
(Stochiometry: 1 : 1)
|
Proteins are synthesised in the cytosol, so they must travel across membranes to reach other cell compartments. Proteins which function in the nucleus pass through the nuclear envelope via the nuclear pores. Small proteins might be capable of diffusing through the pores, although the process is likely to be inefficient. Therefore, almost all well studied nuclear proteins are transported into the nucleus using active translocation through the pore. This requires a targeting motif to bind the transport machinery. Proteins that enter the nucleus in preformed complexes may not require their own targeting motif (Dingwall,1982). Nevertheless, it is clear that most proteins do specify their own import signal and, of these, the vast majority have an NLS that binds to importin-alpha (also termed karyopherin-alpha). Importin-alpha constitutes a multiprotein family in Metazoa with generally similar but perhaps not identical binding specificities (Mason,2009). Importin-alpha is an adaptor protein for importin-beta which interacts with nuclear pore components to effect transport into the nucleus. Several other proteins such as snurportin and transportin may be considered as specialised importin-beta adaptors for specific cargoes like snRNP complexes, while some proteins may interact directly with importin-beta to effect their import (e.g. the viral protein HIV Rev (Henderson,1997)). A striking feature of protein transfer through the nuclear pore is that key roles are played by several linear motifs. Besides the NLS for nuclear import, the CRM1-binding NES motif is found in proteins that are re-exported, while the FG, FxFG and GLFG repeating motifs are found in a subset of nucleoporins (FG-Nups) and are docking sites for the transfer complexes (Terry,2009). The FG-Nups are large, predominantly natively disordered proteins that line the inner pore, extending projections into both the cytosol and nucleoplasm. Importin-beta makes multivalent interactions with motifs in the FG-Nups. Importin-beta stays docked to the FG-Nups while it translocates through the pore. Additional regulatory interactions involving globular interfaces, e.g. with the RAN GTPase, effect allosteric rearrangements of the transport receptors. Overall the nuclear transfer systems are highly cooperative, which is the hallmark of robust cellular operations requiring multiple regulatory inputs to carefully control each step in the process. The "classical" or "conventional" importin-alpha binding NLS is found in the majority of nuclear proteins. It was one of the earliest linear motifs to be described. The dominating feature of the motif is its highly basic nature. It exists in two main variants, a "monopartite" form (mNLS) with a single cluster of basic amino acids, originally found in the sequences of the large T Antigen (SV40) and E1A (Adenovirus) (Kalderon,1984; Smith,1985) and a "bipartite" form (bNLS) with two basic clusters separated by a short linker region, first analysed in Nucleoplasmin (Dingwall,1991). NLS motifs can be found anywhere within a protein sequence provided that the location is well exposed: in practice, they are nearly always in segments of native disorder. Possession of an NLS is sufficient for targeting a protein located in the cytosol into the nucleus. As a consequence, proteins whose localizations are commonly restricted to the cytoplasm can be experimentally directed to the nucleus by fusing an NLS motif onto them. Nuclear import may be very fast but can also be slow, particularly if it is regulated. Thus, the NLS of the Polyomavirus VP1 protein mediated nuclear localization within one minute (Chang,1992). In contrast, for the Adenovirus E1A protein, nuclear localization required up to 30 minutes (Lyons,1987). In nuclear proteins, sequences matching the mNLS are more common than for the bNLS. Structures of NLS peptides in complex with importin-alpha show that the site binding the mNLS also binds the second basic cluster of the bNLS. In the ELM NLS entries, we will refer to this binding site as the "major importin site", in contrast to its interacting counterpart, the "major NLS site" (and similarly to the "minor importin site" and the "minor NLS site" respectively). The NLS itself is known to bind in a predominantly extended conformation (Fontes,2003). Although basic amino acids predominate in the NLS, other amino acids can contribute to the binding interaction, resulting in sufficient variation that accurate definition of motif patterns has not been straightforward. Historically, five positions P1-P5 were used to designate the major importin site (Fontes,2000). Earlier NLS pattern search applications either assumed a fuzzy distribution of the basic residues at these five positions (Nakai,1999) or else used very restricted definitions (Chelsky,1989). Even if it continued to be used for designing pattern search algorithms (Seiler,2006), from a biochemical point of view, such a fuzzy characterization remained unsatisfying. The availability of several crystal structures of NLS peptides in complex with importin-alpha (Conti,2000; Fontes,2003; Tarendeau,2007) has lead to a better understanding, with the positions P2-P5 now designated as the core of the major importin site. The binding interaction is anchored at positions P2 and P3: these are the only positions where basic amino acids are obligatory. In positions P4 and P5, basic residues are preferred to the extent that one of these positions must be Arg or Lys. Pro and hydrophobic residues are reasonable substitutes at P4 and P5. As P4 is the position of the NLS core where most amino acid variation is allowed, we regard [KR][KR]X[KR] to be stronger than [KR][KR][KR]X. Examination of binding peptides, mutational studies and sequence alignments of NLS-containing proteins indicate that adjacent positions modulate the major site by the presence/non-presence of favoured residues: When a hydrophobic residue is found at P4 or P5, favoured residues must be present either in residues preceding P2-P5, or else in positions P6 and P7. As a third option, if these adjacent positions lack favoured residues, then the NLS must be bipartite, with a pair of basic residues occupying the minor site. There are also strongly disfavoured amino acids - in particular the acidic residues (Asp, Glu) appear to be rejected from P1, P4, P5 and P6. In principle, it would be possible to assemble the NLS amino acid preferences into a single regular expression. However, this would be overly complicated and difficult to understand. Therefore for the NLS in ELM, the residue preferences have been split into four patterns, three mNLS and one bNLS. Often a given NLS will match multiple ELM NLS patterns. For proteins that shuttle in and out of the nucleus, it is likely that the NLS can be conditionally inactivated, or they would immediately reimport into the nucleus without executing their cytosolic function. Conditionally inactive NLSes may also be required for the many transcription factors used in transient signalling pathways that are initially targeted to the plasma membrane cytosolic side (Fabbro,2003). Upon cell surface receptor stimulation, they are transferred to the nucleus, where they transiently regulate target genes before being ubiquitinated and destroyed by the proteasome. Such proteins must remain outside the nucleus indefinitely, and therefore the NLS is likely to be activated when the transfer signal is made. Most probably, the main way to control NLS availability is by phosphorylation of the motif at positions that cannot be negatively charged e.g. P1, P4, P5, P6 and possibly at other nearby residue positions (Fontes,2003; Sorokin,2007). |
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement