DOC_CKS1_1
| Accession: | |
|---|---|
| Functional site class: | Cks1 ligand |
| Functional site description: | The cyclin-dependent kinase subunit 1 (Cks1) protein plays a role in cell cycle regulation by binding to and modulating the activity of cyclin-dependent kinases (CDKs). The CDKs in complex with cyclins control progression through the cell cycle by specific phosphorylation of various substrates. Phosphorylation depends on motif-mediated binding of CDK to a phosphorylation site (MOD_CDK_1) and simultaneous binding of the CDK-bound cyclin to the cyclin-recognition site (DOC_CYCLIN_1) on the CDK substrate. These interactions are in many cases sufficient for protein phosphorylation by CDK. However, some substrates additionally interact with cyclin-CDK-associated Cks1, which increases the specificity and efficiency of substrate phosphorylation. This interaction is mediated by phospho-dependent Cks1-binding motifs in the CDK substrates, which allows docking of CDK substrates for phosphorylation and multi-phosphorylation. In addition, Cks1 might also recruit regulators of CDK activity. |
| ELM Description: | The phospho-dependent Cks1-binding motif is actived by priming phosphorylation of the core threonine residue (Koivomagi,2013). Its attached phosphate binds in a cationic pocket on Cks1 while its gamma-methyl group makes additional Van der Waals contacts with Y30 and R42 of Cks1 (4LPA) (McGrath,2013; Arvai,1995). A second conserved motif residue is the proline in the +1 position that specifically binds in a pocket formed by Y30 and T80 of Cks1. Furthermore, this proline is required for the priming phosphorylation of threonine by CDK, which is a proline-directed kinase (MOD_CDK_1). Binding of the motif is further enhanced by a conserved interaction of the L83- and R75-containing pocket on Cks1 with a hydrophobic residue in the -2 position. Peptide arrays showed that aromatic residues are favoured in this position, probably due to R75 making cation-pi contacts (McGrath,2013). These arrays also showed basic residues to be disfavoured in the +2 position. However, multiple sequence alignments using fungal sequences showed a wider preference in the -2 position and a tolerance for arginine in the +2 position. The current motif definition is based on yeast data, as not much experimental evidence is available for vertebrate sequences. Mutational studies showed that the Cks1-docking site is located N-terminally of the secondary phosphorylation site (Koivomagi,2013). In addition to this N-to-C directionality, the phosphorylation rate depends on the distance between the Cks1-docking motif and the target phosphorylation site. Distances less than 10 amino acids were shown to prevent phosphorylation, while distances over 32 amino acids also impaired phosphorylation. These results were confirmed in vivo, showing an optimal distance of 16-20 residues (Koivomagi,2013). This distance dependency indicates that phosphorylation requires simultaneous binding of CDK and Cks1 to the CDK substrate. Several CDK substrates contain multiple Cks1-binding motifs to promote multi-site phosphorylation. |
| Pattern: | [MPVLIFWYQ].(T)P.. |
| Pattern Probability: | 0.0019915 |
| Present in taxon: | Eukaryota |
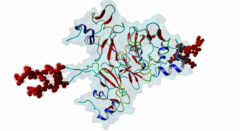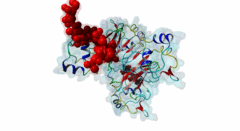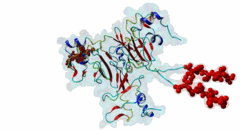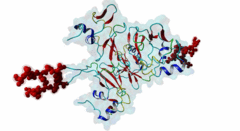
| Interaction Domain: |
CKS (PF01111)
Cyclin-dependent kinase regulatory subunit
(Stochiometry: 1 : 1)
|
The cell division cycle is highly regulated by cyclin-dependent kinases (CDKs) that associate with and are activated by cyclins. Active cyclin-CDK complexes mediate protein phosphorylation, altering the activity and half-life of a wide range of substrates. The substrate specificity of cyclin-CDK complexes is established by the specific recognition of the substrate’s modification site (MOD_CDK_1) by CDK as well as docking of its cyclin-recognition site (DOC_CYCLIN_1) to cyclin. An additional motif-mediated interaction of CDK substrates with the Cyclin-dependent kinases regulatory subunit 1 (Cks1) (1QB3), which can simultaneously bind to CDK and its phosphorylated substrate, allows precise regulation of the binding affinities and multi-phosphorylation of the substrate. Cks1, which was first identified in fission yeast (Hayles,1986), was shown to interact with and activate the G1S-specific Cln2-Cdc28 complex in yeast (Reynard,2000). Absence of Cks1 was shown to prevent both the G1 to S and G2 to M phase transitions (Reynard,2000; Tang,1993; Patra,1996), causing a restricted potential of cell division and influencing cell viability (Hindley,1987). Furthermore, Cks1 is involved in controlling the M to G1 phase transition as depletion of this protein in Xenopus results in a failure to activate the Anaphase Promoting Complex or Cyclosome (APC/C) by CDK-dependent hyper-phosphorylation of the Cdc27 subunit (Patra,1998). Overexpression of Cks1 in fission yeast and Xenopus caused an abnormal and reduced cell cycle with an elongated G2 phase and a decreased Cyclin B-Cdc2 activation, indicating that Cks1 functions both as a positive and negative regulator of the cell cycle (Hayles,2010; Patra,1996). In mammals, two Cks1 paralogs (Cks1 and Cks2) with high sequence similarity are expressed. They appear to have overlapping functions as a knockout of either does not affect viability, while a double knockout is lethal in mice (Martinsson-Ahlzen,2008). Cks1 was shown to interact with both CDK1 (Draetta,1987) and CDK2 in higher eukaryotes (1BUH) (Bourne,1996; Zhang,1995). Similar to yeast and Xenopus, depletion of Cks1 in mouse embryonic fibroblasts causes cell cycle arrest in the G2 phase, and this effect was likely related to a decrease in Cyclin B1, Cyclin A, and CDK1 expression (Martinsson-Ahlzen,2008). These results show that Cks1 regulates several steps of the cell cycle and plays a prominent role in cell cycle phase transitions. An important role of Cks1 in cell cycle regulation is to increase the specificity and efficiency of CDK-catalysed phosphorylation by providing an additional docking site for primed CDK substrates. Specific binding to Cks1 is mediated by the phospho-dependent Cks1-binding motif present in several CDK substrates. This motif forms a multipartite binding site with the CDK modification motif (MOD_CDK_1) and the cyclin-binding docking motif (DOC_CYCLIN_1). The crystal structure of the Cks1-CDK2 complex further shows that the catalytic domain of CDK2 and the motif-binding domain of Cks1 are located in spatial proximity, indicating that during binding the CDK modification site and the Cks1-binding motif on the substrate are located in spatial proximity (1BUH) (Bourne,1996). However, Cks1 might also provide a docking site for regulators of CDK activity such as the yeast Swe1 kinase (McGrath,2013). In addition, Cks1, in cooperation with the F box protein Skp2, also controls CDK activity by mediating the degradation of CDK inhibitors such as p27Kip1 through phospho-dependent recognition of a specific degron (DEG_SCF_SKP2-CKS1_1) in these inhibitors (Hao,2005). |
-
Loss of cks1 homeostasis deregulates cell division cycle.
Krishnan A, Nair SA, Pillai MR
J Cell Mol Med 2010 Apr 19; 14 (1-2), 154-64
PMID: 19228269
-
Multisite phosphorylation networks as signal processors for Cdk1.
Koivomagi M, Ord M, Iofik A, Valk E, Venta R, Faustova I, Kivi R, Balog ER, Rubin SM, Loog M
Nat Struct Mol Biol 2013 Dec 05; 20 (12), 1415-24
PMID: 24186061
-
Cks confers specificity to phosphorylation-dependent CDK signaling pathways.
McGrath DA, Balog ER, Koivomagi M, Lucena R, Mai MV, Hirschi A, Kellogg DR, Loog M, Rubin SM
Nat Struct Mol Biol 2013 Dec 05; 20 (12), 1407-14
PMID: 24186063
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement