DEG_SIAH_1
| Accession: | |
|---|---|
| Functional site class: | Siah binding Motif |
| Functional site description: | The members of the SINA/Siah family have domain architecture consisting of a RING domain, two zinc finger motifs and a substrate/adaptor binding domain (SBD) that mediates the interaction with the binding partners. |
| ELM Description: | The PxAxVxP motif was first described to confer a high-affinity binding to the Siah/PHYL interaction. It has been detected in other Siah interacting proteins, including DCC, KLF10, OBF-1. The motif binds by partial Beta-augmentation. The present pattern is stricter than proposed by House,2006. It will find most verified instances but may miss some candidates. |
| Pattern: | .P.A.V.P[^P] |
| Pattern Probability: | 0.0000271 |
| Present in taxon: | Metazoa |
| Interaction Domain: |
Sina (PF03145)
Seven in absentia protein family
(Stochiometry: 1 : 1)
|
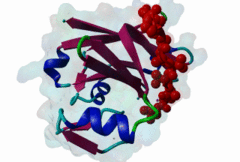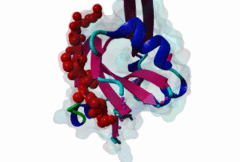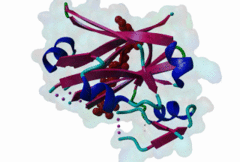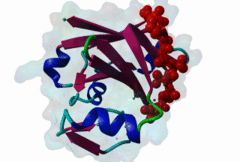
Proteasomal degradation of proteins plays important roles in cell physiology. In eukaryotic cells, this process involves the specific covalent modification by a highly conserved small regulatory protein, ubiquitin, which labels target proteins for proteolysis and subsequent degradation. Ubiquitination reaction is carried out by the E1 (ubiquitin activating proteins) - E2 (ubiquitin conjugating proteins) - E3 (ubiquitin ligases) cascade of enzymes. The specificity for targeting substrates for ubiquitination is mainly conferred by the E3 proteins. Siah proteins are members of an evolutionarily highly conserved family of E3 ubiquitin ligases which regulate the ubiquitination and proteasome-dependent degradation of several proteins. The mammalian Siah genes are homologues of the Drosophila SINA (seven in absentia homologue), which is required for the R7 cell determination in the developing eyes, and they are characterized by the presence of a RING finger domain that is responsible for E3 activity. SIAH/sina proteins interact with several proteins and participate in the regulation of some of them through ubiquitination and proteasome-dependent degradation. Degraded proteins include Drosophila repressor Tramtrack (Ttk88) (Li,1997; Tang,1997), netrin-1 receptor deleted in colorectal cancer (DCC) (Hu,1997), coactivator BOB.1/OBF.1 (Boehm,2001; Tiedt,2001), nuclear receptor corepressor (NcoR) (Zhang,1998), the transcriptional repressor TIEG-1 (Johnsen,2002) and Kid (Germani,2001), a protein necessary for chromosome movement during mitosis and meiosis. SIAH-1 also interacts with a complex including step1, Ebi, Sip (SIAH-interacting protein) and APC (adenomatosis polyposis colonic protein), which facilitates the degradation of beta-catenin in a p53-dependent manner (Liu,2001; Matsuzawa,2001). The C-terminus region of the Siah proteins has been demonstrated to be a substrate- and cofactor- interaction module (Substrate-Binding Domain, SBD. The X-ray analysis of the Siah SBD revealed an eight-stranded beta sandwich fold (Polekhina,2002). House and colleagues (House,2003) have identified a binding motif PxAxVxP (named Siah degron), present in the Drosophila protein PHYL, which binds with high affinity to the SINA and Siah SBDs. The degron motif is present in several Siah degradation targets. The structure of the Siah1 in complex with a degron-containing peptide from the adaptor protein Phyllopod has been recently solved (2AN6) (House,2006). Other Siah-interacting proteins, including KID, APC, synphylin-1, carry similar peptides that do not perfectly match the reported consensus degron motif. |
-
Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin
degradation linked to p53 responses.
Matsuzawa SI, Reed JC
Mol Cell 2001 May; 7 (5), 915-26
PMID: 11389839
-
Siah-1 mediates a novel beta-catenin degradation pathway linking p53 to
the adenomatous polyposis coli protein.
Liu J, Stevens J, Rote CA, Yost HJ, Hu Y, Neufeld KL, White RL, Matsunami N
Mol Cell 2001 May; 7 (5), 927-36
PMID: 11389840
-
Siah ubiquitin ligase is structurally related to TRAF and modulates
TNF-alpha signaling.
Polekhina G, House CM, Traficante N, Mackay JP, Relaix F, Sassoon DA, Parker MW, Bowtell DD
Nat Struct Biol 2002 Jan; 9 (1), 68-75
PMID: 11742346
-
A binding motif for Siah ubiquitin ligase.
House CM, Frew IJ, Huang HL, Wiche G, Traficante N, Nice E, Catimel B, Bowtell DD
Proc Natl Acad Sci U S A 2003 Mar 18; 100 (6), 3101-6
PMID: 12626763
-
I Siah substrate!
Briant DJ, Ceccarelli DF, Sicheri F
Structure 2006 Apr; 14 (4), 627-8
PMID: 16615900
-
Elucidation of the substrate binding site of Siah ubiquitin ligase.
House CM, Hancock NC, Moller A, Cromer BA, Fedorov V, Bowtell DD, Parker MW, Polekhina G
Structure 2006 Apr; 14 (4), 695-701
PMID: 16615911
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
| Acc., Gene-, Name | Start | End | Subsequence | Logic | #Ev. | Organism | Notes |
|---|---|---|---|---|---|---|---|
| Q13118 KLF10 KLF10_HUMAN |
200 | 208 | AARKNIPCAAVSPNRSKCER | TP | 1 | Homo sapiens (Human) | |
| Q27934 phyl PHYL_DROME |
115 | 123 | ERTKLRPVAMVRPTVRVQPQ | TP | 5 | Drosophila melanogaster (Fruit fly) | |
| Q7Z6J0 SH3RF1 SH3R1_HUMAN |
600 | 608 | AHNQERPTAAVTPIQVQNAA | TP | 2 | Homo sapiens (Human) | |
| Q16633 POU2AF1 OBF1_HUMAN |
46 | 54 | SGAAPAPTAVVLPHQPLATY | TP | 2 | Homo sapiens (Human) | |
| Q86TG7 PEG10 PEG10_HUMAN |
253 | 261 | RKPRSPPRALVLPHIASHHQ | TP | 2 | Homo sapiens (Human) | |
| O75553 DAB1 DAB1_HUMAN |
360 | 368 | VMGAQPPVAQVMPGAQPIAW | TP | 2 | Homo sapiens (Human) | |
| Q9HB71 CACYBP CYBP_HUMAN |
59 | 67 | LLDNEKPAAVVAPITTGYTV | TP | 4 | Homo sapiens (Human) | |
| Q9UHB7 AFF4 AFF4_HUMAN |
252 | 260 | NSMLQKPTAYVRPMDGQESM | TP | 2 | Homo sapiens (Human) | |
| P43146 DCC DCC_HUMAN |
1331 | 1339 | APSRTIPTACVRPTHPLRSF | TP | 1 | Homo sapiens (Human) |
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement