LIG_EABR_CEP55_1
| Accession: | |
|---|---|
| Functional site class: | Cep55 EABR-binding motif |
| Functional site description: | The Cep55-binding motif mediates binding of proteins to the EABR domain of Cep55 and thereby recruits these proteins to the midbody, where they are involved in somatic cell abscission or formation of intercellular bridges between differentiating germ cells. |
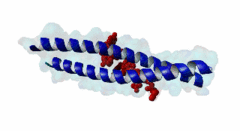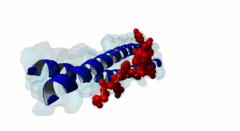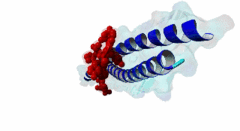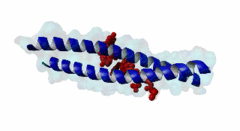
| ELM Description: | The LIG_EABR_CEP55_1 motif binds to the Cep55 EABR domains, which form a non-canonical dimeric parallel coiled-coil. One Cep55 dimer binds a single copy of the motif (3E1R). In the central coiled-coil regions, non-canonical interactions between the two coils cause them to be pushed apart and the structure to become asymmetric, which results in the formation of the motif-binding site. The two motif regions that were found to be essential for binding are the GPP sequence at position 4 to 6 and the tyrosine residue in position 10. The GPP sequence makes extensive contacts with different residues of Cep55, including Trp184 and Tyr187, whose mutation disrupts binding of the Alix peptide. The Pro in position 5 makes the most extensive contacts of the three residues in the GPP sequence. The Alix GPP to AAA triple mutant peptide does not bind, while individual mutations show that Gly in position 4 (3-fold reduction), Pro in position 5 (60-fold reduction) and Pro in position 6 (17-fold reduction) make different but significant contributions to the interaction (Lee,2008). Similar results were obtained for Tex14 (Iwamori,2010). Also for Tsg101, mutation of both prolines abolishes binding (Morita,2007). In addition, there is a conserved Ala in position 2. A triple ATG (position 2 to 4 of the motif) to GAA mutant of Tsg101 fails to bind Cep55, indicating that this Ala is also important (Morita,2007). Mutation of the second major interaction site in the motif, the conserved Tyr residue in position 10, also abolishes binding of Alix (Lee,2008) and Tex14 (Iwamori,2010) and reduces binding of Tsg101 (Morita,2007), indicating its importance for binding specificity. Downstream of this Tyr there is a preference for hydrophobic and proline residues, but whether they are important for this motif or belong to overlapping motifs is not known. |
| Pattern: | .A.GPP.{2,3}Y. |
| Pattern Probability: | 0.0000014 |
| Present in taxon: | Metazoa |
| Interaction Domain: |
EABR (PF12180)
TSG101 and ALIX binding domain of CEP55
(Stochiometry: 2 : 1)
|
The endosomal sorting complex required for transport (ESCRT) pathway plays an important role in different cellular processes that depend on membrane fission, including release of viral particles, formation of late endosomal multivesicular bodies (MVBs) and abscission of daughter cells upon completion of cytokinesis (McCullough,2013). The ESCRT machinery consists of five distinct multisubunit complexes: ESCRT-0, -I, -II, -III, and the vacuolar protein sorting-associated protein 4 (Vps4) complexes. Each of these has a specific function in mediating membrane scission, and this modularity allows recruitment of the ESCRT machinery to different cellular locations and its integration in different cellular processes by means of different adaptor proteins. While the early acting ESCRT-0, -I and -II complexes mediate ESCRT assembly and membrane remodeling, the late acting ESCRT-III and Vps4 complexes are responsible for the membrane scission activity and disassembly of the machinery, respectively (Schmidt,2012). The midbody protein Cep55 (Centrosomal protein of 55kDa) is the adaptor protein that recruits the ESCRT machinery to the midbody region during cytokinesis to enable fission of the plasma membrane between dividing cells. Cep55 homodimers directly interact via their EABR domains (PF12180) with the LIG_EABR_CEP55_1 motifs in PDCD6IP/Alix and the ESCRT-I subunit Tsg101, which in turn recruit components of the ESCRT-III complex and are thus essential for proper cell abscission (Caballe,2011). In contrast to somatic cells, differentiating germ cells do not complete cytokinesis but remain connected through intercellular bridges that are essential for proper gametogenesis and fertility. These stable structures are composed of general cytokinesis-related components and additional germ cell-specific factors like Tex14 (Greenbaum,2011). Tex14 negatively regulates abscission by competitively binding to Cep55 via its LIG_EABR_CEP55_1 motif, thus preventing recruitment of Alix and Tsg101 and subsequent membrane fission (Iwamori,2010). |
-
Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis.
Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI
EMBO J 2007 Oct 03; 26 (19), 4215-27
PMID: 17853893
-
Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55.
Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH
Science 2008 Oct 24; 322 (5901), 576-80
PMID: 18948538
-
TEX14 interacts with CEP55 to block cell abscission.
Iwamori T, Iwamori N, Ma L, Edson MA, Greenbaum MP, Matzuk MM
Mol Cell Biol 2010 Apr 08; 30 (9), 2280-92
PMID: 20176808
-
Germ cell intercellular bridges.
Greenbaum MP, Iwamori T, Buchold GM, Matzuk MM
Cold Spring Harb Perspect Biol 2011 Aug 02; 3 (8), a005850
PMID: 21669984
-
ESCRT machinery and cytokinesis: the road to daughter cell separation.
Caballe A, Martin-Serrano J
Traffic 2011 Sep 12; 12 (10), 1318-26
PMID: 21722282
-
Membrane Fission Reactions of the Mammalian ESCRT Pathway.
McCullough J, Colf LA, Sundquist WI
Annu Rev Biochem 2013 Mar 26; 0 (0), 0
PMID: 23527693
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
| Acc., Gene-, Name | Start | End | Subsequence | Logic | #Ev. | Organism | Notes |
|---|---|---|---|---|---|---|---|
| Q99816 TSG101 TS101_HUMAN |
154 | 164 | YPPYQATGPPNTSYMPGMPG | TP | 9 | Homo sapiens (Human) | |
| Q61187 Tsg101 TS101_MOUSE |
155 | 165 | YPPYTATGPPNTSYMPGMPS | TP | 2 | Mus musculus (House mouse) | |
| Q8WUM4 PDCD6IP PDC6I_HUMAN |
797 | 807 | APPPQAQGPPYPTYPGYPGY | TP | 15 | Homo sapiens (Human) | |
| Q9WU78 Pdcd6ip PDC6I_MOUSE |
798 | 808 | APPPQAQGPPYPTYPGYPGY | TP | 2 | Mus musculus (House mouse) | |
| Q8IWB6 TEX14 TEX14_HUMAN |
792 | 802 | YKLPLAVGPPSLNYIPPVLQ | TP | 4 | Homo sapiens (Human) | |
| Q7M6U3 Tex14 TEX14_MOUSE |
788 | 798 | AKFQPAVGPPSLAYLPPVMQ | TP | 5 | Mus musculus (House mouse) |
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement