LIG_GYF
| Accession: | |
|---|---|
| Functional site class: | GYF ligand |
| Functional site description: | Proline-rich sequences recognized by GYF domains - involved in lymphoid signaling |
| ELM Description: | LIG_GYF is a proline-rich sequence specifically recognized by GYF domains. Known cases so far involve the CD2 binding protein CD2BP2, which specifically recognizes the cytoplasmic tail of CD2 surface receptor. The LIG_GYF sequence is generally composed of a pair of similar proline-rich sequences and has been shown to compete in vitro with the SH3 domain of the src-family FYN kinase. Each proline-rich sequence is spanned by positively charged residues that have complementary binding partners on the GYF domain. Due to the limited experimental data available, the current pattern is certainly overdetermined and a better description should be possible in future. |
| Pattern: | [QHR].{0,1}P[PL]PP[GS]H[RH] |
| Pattern Probability: | 1.686e-08 |
| Present in taxons: | Eukaryota Homo sapiens Mus musculus Rattus norvegicus |
| Interaction Domain: |
GYF (PF02213)
GYF domain
(Stochiometry: 1 : 1)
|
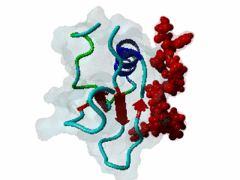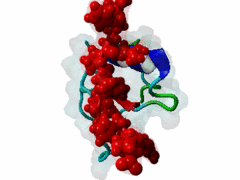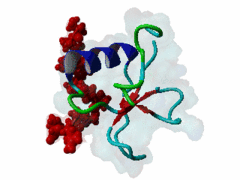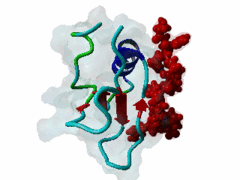
The glycine-tyrosine-phenylalanine, or GYF, domain has been identified at the N-term of a number of proteins in organisms ranging from S.pombe to H.sapiens with fairly conserved sequence. It interacts with proline-rich sequences and, in humans, it has been shown to recognize specifically two PPPPGHR repeats at the C-term of the CD2 cell surface receptor. Despite functioning as a proline-rich peptide binding domain, the GYF fold is structurally unrelated to the SH3 or WW domains. The NMR structure of the GYF domain of the CD2BP2 protein has been solved in complex with a proline-rich peptide (1L2Z) and the residues of the domain involved in binding have been identified. The binding pocket is very hydrophobic with a relatively smooth, concave surface. The GYF tripeptide (conserved at the C term of the domain sequence) is involved in the binding. |
-
The GYF domain is a novel structural fold that is involved in lymphoid
signaling through proline-rich sequences.
Freund C, Dotsch V, Nishizawa K, Reinherz EL, Wagner G
Nat Struct Biol 1999 Jul; 6 (7), 656-60
PMID: 10404223
-
Dynamic interaction of CD2 with the GYF and the SH3 domain of
compartmentalized effector molecules.
Freund C, Kuhne R, Yang H, Park S, Reinherz EL, Wagner G
EMBO J 2002 Nov 15; 21 (22), 5985-95
PMID: 12426371
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
| Acc., Gene-, Name | Start | End | Subsequence | Logic | #Ev. | Organism | Notes |
|---|---|---|---|---|---|---|---|
| P06729 CD2 CD2_HUMAN |
295 | 303 | SQAPSHRPPPPGHRVQHQPQ | TP | 4 | Homo sapiens (Human) | |
| P06729 CD2 CD2_HUMAN |
281 | 289 | PATSQHPPPPPGHRSQAPSH | TP | 2 | Homo sapiens (Human) | |
| P37998 CD2 CD2_HORSE |
276 | 284 | NPAASQPPPPPSHRPQAPGH | TP | 1 | Equus caballus (Horse) |
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement