| Accession | Acc. Gene-, Name | Start | End | Subsequence | Logic | PDB | Organism | Length |
|---|---|---|---|---|---|---|---|---|
| ELMI001607 | Q99700 ATXN2 ATX2_HUMAN |
912 | 924 | VRKSTLNPNAKEFNPRSFSQ | TP |
3KTR 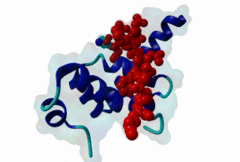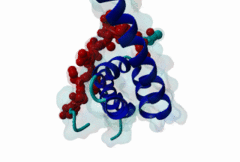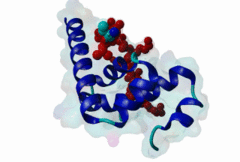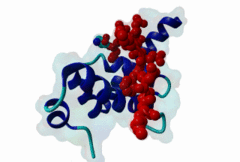
|
Homo sapiens (Human) | 1313 |
![]() Instance evidence
Instance evidence
| Evidence class | PSI-MI | Method | BioSource | PubMed | Logic | Reliability | Notes |
|---|---|---|---|---|---|---|---|
| experimental | MI:0065 | isothermal titration calorimetry | in vitro | Lim,2006 | support | likely | InteractionDetection |
| experimental | MI:0077 | nuclear magnetic resonance | in vitro | Lim,2006 | support | likely | InteractionDetection FeatureDetection |
| experimental | MI:0107 | surface plasmon resonance | in vitro | Lim,2006 | support | certain | InteractionDetection FeatureDetection |
| experimental | MI:0065 | isothermal titration calorimetry | in vitro | Lim,2006 | support | certain | InteractionDetection |
| experimental | MI:0114 | x-ray crystallography | in vitro | Kozlov,2010 | support | certain | InteractionDetection FeatureDetection |
![]() Interactions
Interactions
| Uniprot Id | Domain family | Domain Start | Domain End | Affinity Min/Max (µMol) | Notes |
|---|---|---|---|---|---|
| (P11940) PABP1_HUMAN | PF00658
(PABP)
Poly-adenylate binding protein, unique domain |
543 | 614 | 0.68/0.76 | [mitab][xml] |
| (Q62671) UBR5_RAT | PF00658
(PABP)
Poly-adenylate binding protein, unique domain |
499 | 571 | 9.9/9.9 | [mitab][xml] |
![]() Pathways
Pathways
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
