| Accession | Acc. Gene-, Name | Start | End | Subsequence | Logic | PDB | Organism | Length |
|---|---|---|---|---|---|---|---|---|
| ELMI001555 | P05067 APP A4_HUMAN |
756 | 763 | SKMQQNGYENPTYKFFEQMQ | TP |
1AQC 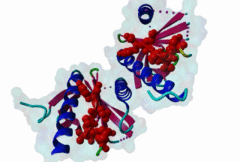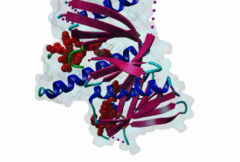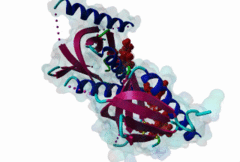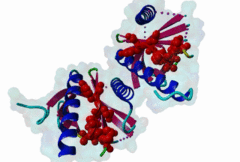 1X11
1X11  3DXC
3DXC  3DXD
3DXD  3DXE
3DXE 
|
Homo sapiens (Human) | 770 |
![]() Instance evidence
Instance evidence
| Evidence class | PSI-MI | Method | BioSource | PubMed | Logic | Reliability | Notes |
|---|---|---|---|---|---|---|---|
| experimental | MI:0096 | pull down | in vivo/in vitro | support | certain | InteractionDetection | |
| experimental | MI:0519 | glutathione s tranferase tag | in vitro | Howell,1999 | support | certain | |
| experimental | MI:0074 | mutation analysis | in vivo/in vitro | Howell,1999 | support | certain | FeatureDetection |
| experimental | MI:0018 | two hybrid | in vivo | Howell,1999 | support | certain | InteractionDetection |
| experimental | MI:0107 | surface plasmon resonance | in vitro | Zhang,1997 | support | certain | InteractionDetection FeatureDetection |
| experimental | MI:0114 | x-ray crystallography | in vitro | Zhang,1997 | support | certain | InteractionDetection FeatureDetection |
| experimental | MI:0065 | isothermal titration calorimetry | in vitro | Radzimanowski,2008 | support | certain | InteractionDetection |
| experimental | MI:0114 | x-ray crystallography | in vitro | Radzimanowski,2008 | support | certain | InteractionDetection FeatureDetection |
| experimental | MI:0005 | alanine scanning | in vivo/in vitro/in silico | Borg,1996 | support | certain | FeatureDetection |
![]() Interactions
Interactions
| Uniprot Id | Domain family | Domain Start | Domain End | Affinity Min/Max (µMol) | Notes |
|---|---|---|---|---|---|
| (Q02410) APBA1_HUMAN | PF00640
(PID)
Phosphotyrosine interaction domain (PTB/PID) |
461 | 618 | 0.32/0.32 | [mitab][xml] |
| (O00213) APBB1_HUMAN | PF00640
(PID)
Phosphotyrosine interaction domain (PTB/PID) |
0.2/0.2 | [mitab][xml] | ||
| (O75553) DAB1_HUMAN | PF00640
(PID)
Phosphotyrosine interaction domain (PTB/PID) |
42 | 168 | 0.55/0.55 | [mitab][xml] |
![]() Switches
Switches
This ELM instance is part of the following 2 switching mechanisms annotated at the switches.ELM resource:
-
SWTI000201:
Phosphorylation of T743 adjacent to the PTB-binding motif of Amyloid beta A4 protein (APP) reduces the affinity for Amyloid beta A4 precursor protein-binding family B member 1 (APBB1).
-
SWTI000292:
Phosphorylation of Y757 in APP (Amyloid beta A4 protein (APP)) switches its specificity from PTB domain containing proteins, like Amyloid beta A4 precursor protein-binding family B member 1 (APBB1), which is involved in trafficking and processing of APP, to SH2 domain containing proteins, such as Growth factor receptor-bound protein 2 (GRB2).
![]() Pathways
Pathways
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement


