| Accession | Acc. Gene-, Name | Start | End | Subsequence | Logic | PDB | Organism | Length |
|---|---|---|---|---|---|---|---|---|
| ELMI001475 | P03347 gag GAG_HV1B1 |
495 | 504 | PIDKELYPLTSLRSLFGNDP | TP |
2R02 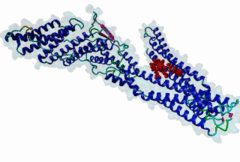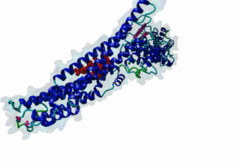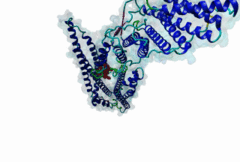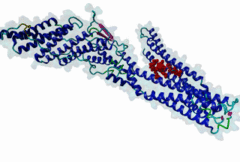
|
Human immunodeficiency virus type 1 BH10 | 512 |
![]() Instance evidence
Instance evidence
| Evidence class | PSI-MI | Method | BioSource | PubMed | Logic | Reliability | Notes |
|---|---|---|---|---|---|---|---|
| experimental | MI:0096 | pull down | in vivo/in vitro | support | certain | InteractionDetection | |
| experimental | MI:0519 | glutathione s tranferase tag | in vitro | Strack,2003 | support | certain | |
| experimental | MI:0107 | surface plasmon resonance | in vitro | Fisher,2007 | support | certain | InteractionDetection FeatureDetection |
| experimental | MI:0018 | two hybrid | in vivo | Martin-Serrano,2003 | support | certain | InteractionDetection |
| experimental | MI:0065 | isothermal titration calorimetry | in vitro | Lee,2007 | support | certain | InteractionDetection |
| experimental | MI:0107 | surface plasmon resonance | in vitro | Zhai,2008 | support | certain | InteractionDetection FeatureDetection |
| experimental | MI:0114 | x-ray crystallography | in vitro | Zhai,2008 | support | certain | InteractionDetection FeatureDetection |
![]() Interactions
Interactions
| Uniprot Id | Domain family | Domain Start | Domain End | Affinity Min/Max (µMol) | Notes |
|---|---|---|---|---|---|
| (Q8WUM4) PDC6I_HUMAN | PF13949
(ALIX_LYPXL_bnd)
ALIX V-shaped domain binding to HIV |
408 | 702 | [mitab][xml] |
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
