| Accession | Acc. Gene-, Name | Start | End | Subsequence | Logic | PDB | Organism | Length |
|---|---|---|---|---|---|---|---|---|
| ELMI001665 | Q8K4J6 Mkl1 MKL1_MOUSE |
61 | 80 | LKRKIRSRPERAELVRMHIL | TP |
2V52 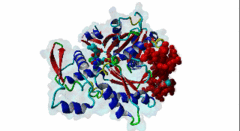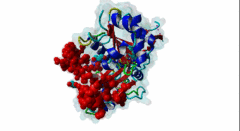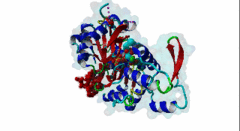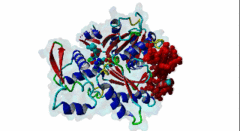 2YJE
2YJE  2YJF
2YJF 
|
Mus musculus (House mouse) | 964 |
![]() Instance evidence
Instance evidence
| Evidence class | PSI-MI | Method | BioSource | PubMed | Logic | Reliability | Notes |
|---|---|---|---|---|---|---|---|
| experimental | MI:0096 | pull down | in vivo/in vitro | support | certain | InteractionDetection | |
| experimental | MI:0519 | glutathione s tranferase tag | in vitro | Vartiainen,2007 | support | certain | |
| experimental | MI:0096 | pull down | in vivo/in vitro | support | certain | InteractionDetection | |
| experimental | MI:0519 | glutathione s tranferase tag | in vitro | Guettler,2008 | support | certain | |
| experimental | MI:0074 | mutation analysis | in vivo/in vitro | Guettler,2008 | support | certain | FeatureDetection |
| experimental | MI:0053 | fluorescence polarization spectroscopy | in vitro | Guettler,2008 | support | certain | InteractionDetection |
| experimental | MI:0367 | green fluorescent protein tag | in vivo | Guettler,2008 | support | certain | |
| experimental | MI:0005 | alanine scanning | in vivo/in vitro/in silico | Guettler,2008 | support | certain | FeatureDetection |
| experimental | MI:0105 | structure based prediction | in silico | Mouilleron,2011 | support | certain | InteractionDetection |
| experimental | MI:0074 | mutation analysis | in vivo/in vitro | Mouilleron,2011 | support | certain | FeatureDetection |
| experimental | MI:0071 | molecular sieving | in vivo/in vitro | Mouilleron,2011 | support | certain | InteractionDetection |
| experimental | MI:0016 | circular dichroism | in vitro | Mouilleron,2011 | support | certain | InteractionDetection |
| experimental | MI:0005 | alanine scanning | in vivo/in vitro/in silico | Mouilleron,2011 | support | certain | FeatureDetection |
| experimental | MI:0403 | colocalization | in vitro | Pawlowski,2010 | support | certain | |
| experimental | MI:0005 | alanine scanning | in vivo/in vitro/in silico | Pawlowski,2010 | support | certain | FeatureDetection |
| experimental | MI:0074 | mutation analysis | in vivo/in vitro | Pawlowski,2010 | support | certain | FeatureDetection |
| experimental | MI:0071 | molecular sieving | in vivo/in vitro | Vartiainen,2007 | support | certain | InteractionDetection |
| experimental | MI:0005 | alanine scanning | in vivo/in vitro/in silico | Vartiainen,2007 | support | certain | FeatureDetection |
| experimental | MI:0074 | mutation analysis | in vivo/in vitro | Vartiainen,2007 | support | certain | FeatureDetection |
| experimental | MI:0074 | mutation analysis | in vivo/in vitro | Miralles,2003 | support | certain | FeatureDetection |
| experimental | MI:0019 | coimmunoprecipitation | in vivo/in vitro | Miralles,2003 | support | certain | InteractionDetection |
| experimental | MI:0403 | colocalization | in vitro | Miralles,2003 | support | certain | |
| experimental | MI:0101 | sequence based prediction | in silico | Miralles,2003 | support | certain | InteractionDetection |
| experimental | MI:0074 | mutation analysis | in vivo/in vitro | Mouilleron,2008 | support | certain | FeatureDetection |
| experimental | MI:0053 | fluorescence polarization spectroscopy | in vitro | Mouilleron,2008 | support | certain | InteractionDetection |
| experimental | MI:0005 | alanine scanning | in vivo/in vitro/in silico | Mouilleron,2008 | support | certain | FeatureDetection |
| experimental | MI:0105 | structure based prediction | in silico | Mouilleron,2008 | support | certain | InteractionDetection |
![]() Interactions
Interactions
| Uniprot Id | Domain family | Domain Start | Domain End | Affinity Min/Max (µMol) | Notes |
|---|---|---|---|---|---|
| (P68135) ACTS_RABIT | PF00022
(Actin)
Actin |
4 | 377 | 2.1/2.5 | [mitab][xml] |
![]() Switches
Switches
This ELM instance is part of the following 1 switching mechanism annotated at the switches.ELM resource:
-
SWTI000318:
Hiding of the NLS of MKL/myocardin-like protein 1 (Mkl1) by binding of G-actin to the RPEL motifs of MKL/myocardin-like protein 1 (Mkl1) prevents translocation of this transcription factor to the nucleus.
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement

