| Accession | Acc. Gene-, Name | Start | End | Subsequence | Logic | PDB | Organism | Length |
|---|---|---|---|---|---|---|---|---|
| ELMI001385 | P24864 CCNE1 CCNE1_HUMAN |
393 | 399 | SPLPSGLLTPPQSGKKQSSG | TP |
2OVQ 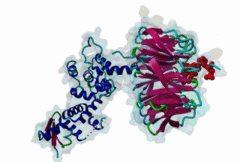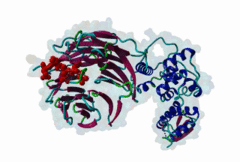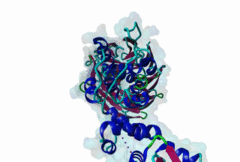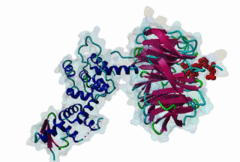
|
Homo sapiens (Human) | 410 |
![]() Instance evidence
Instance evidence
| Evidence class | PSI-MI | Method | BioSource | PubMed | Logic | Reliability | Notes |
|---|---|---|---|---|---|---|---|
| experimental | MI:0114 | x-ray crystallography | in vitro | Hao,2007 | support | certain | InteractionDetection FeatureDetection |
| experimental | MI:0065 | isothermal titration calorimetry | in vitro | Hao,2007 | support | certain | InteractionDetection |
| experimental | MI:0074 | mutation analysis | in vivo/in vitro | Welcker,2003 | support | certain | FeatureDetection |
![]() Switches
Switches
This ELM instance is part of the following 1 switching mechanism annotated at the switches.ELM resource:
-
SWTI000294:
Phosphorylation of G1/S-specific cyclin-E1 (CCNE1) at S399 generates a docking site for Glycogen synthase kinase-3 beta (GSK3B). Subsequent phosphorylation of CCNE1 by Glycogen synthase kinase-3 beta (GSK3B) at T395 switches the specificity of CCNE1 to the F-box/WD repeat-containing protein 7 (FBXW7), which recruits CCNE1 to the SCF ubiquitin ligase complex to mark CCNE1 for degradation.
![]() Pathways
Pathways
- Cell cycle
- Cellular senescence
- Cushing syndrome
- Epstein Barr virus infection
- Gastric cancer
- Hepatitis B
- Human T cell leukemia virus 1 infection
- Human papillomavirus infection
- Measles
- MicroRNAs in cancer
- Oocyte meiosis
- PI3K Akt signaling pathway
- Pathways in cancer
- Prostate cancer
- Small cell lung cancer
- Viral carcinogenesis
- p53 signaling pathway
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement

